Note
Click here to download the full example code
2.6.8.22. Watershed segmentation¶
This example shows how to do segmentation with watershed.
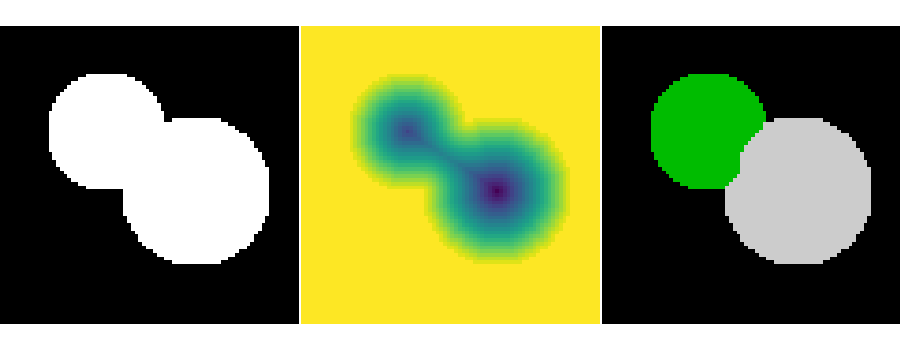
import numpy as np
from skimage.morphology import watershed
from skimage.feature import peak_local_max
import matplotlib.pyplot as plt
from scipy import ndimage
# Generate an initial image with two overlapping circles
x, y = np.indices((80, 80))
x1, y1, x2, y2 = 28, 28, 44, 52
r1, r2 = 16, 20
mask_circle1 = (x - x1) ** 2 + (y - y1) ** 2 < r1 ** 2
mask_circle2 = (x - x2) ** 2 + (y - y2) ** 2 < r2 ** 2
image = np.logical_or(mask_circle1, mask_circle2)
# Now we want to separate the two objects in image
# Generate the markers as local maxima of the distance
# to the background
distance = ndimage.distance_transform_edt(image)
local_maxi = peak_local_max(
distance, indices=False, footprint=np.ones((3, 3)), labels=image)
markers = ndimage.label(local_maxi)[0]
labels = watershed(-distance, markers, mask=image)
plt.figure(figsize=(9, 3.5))
plt.subplot(131)
plt.imshow(image, cmap='gray', interpolation='nearest')
plt.axis('off')
plt.subplot(132)
plt.imshow(-distance, interpolation='nearest')
plt.axis('off')
plt.subplot(133)
plt.imshow(labels, cmap='nipy_spectral', interpolation='nearest')
plt.axis('off')
plt.subplots_adjust(hspace=0.01, wspace=0.01, top=1, bottom=0, left=0,
right=1)
plt.show()
Total running time of the script: ( 0 minutes 0.055 seconds)
